What is Amplification Refractory Mutation System?
Background
Amplification Refractory Mutation System (ARMS), also called allele-specific polymerase chain reaction (ASP) and polymerase chain reaction amplification of specific alleles (PASA), is a method used to detect single base pair mutations. The PCR-based technique can be used to analyze a wide variety of germ-line and somatic mutations, such as sickle-cell anemia. ARMS has the ability to isolate low levels of a mutant sequence in a background of wild-type DNA. The system depends on the specificity of a primer for the normal sequence and another primer for the mutation.
The first demonstration of ARMS occurred in 1989 by Newton et al. The system, when performed correctly, is simple, reliable, and is able to show the genotype for a given allele. In the study, ARMS was applied to patients with α1-antitrypsin deficiency, who were either carriers of the disease and to non-affected individuals. The findings of the first study were in full agreement with allele assignments that were derived from direct PCR sequencing.
Process
As previously mentioned, ARMS is noted for being highly specific for a normal primer and a mutant primer. ARMS is based on allele-specific priming of the PCR process which, after the process is complete, can reveal the genotype of the patient in question. The process requires normal primers, common primers and mutant primers. The normal primer is matched at its 3’ end to the normal nucleotide and is mismatched at its 3’ end to the mutant nucleotide. The reverse happens as well; the mutant primer is matched at the mutant nucleotide and mismatched at the normal sequence.
A mismatched 3’ end of a the primer prevents nucleotide expansion during DNA synthesis in a PCR reaction, and, consequently, no reaction occurs. In an ARMS reaction, DNA is split into two aliquots, one undergoes reaction with normal primers, while the other with mutant primers. The formation of a product is measured by gel electorphoresis. If a product is formed when the normal primers are used, then the DNA sample used came from a patient who is homozygous for the normal nucleotide. If products are present in both reactions, the patient is heterozygous. Lastly, if only one PCR product exists from the mutant primers, then the patient is homozygous for the mutation.
Because of ARMS’ ability to detect single nucleotide polymorphisms (SNPs) in heterozygote patients, the technique has been increasingly popular. A few studies have been produced that examine mutations in patients where ARMS has been used.
Detection of JAK2 V617F Mutation in Human Myeloproliferative Disorders
A mutation in JAK2 has been discovered in human myeloproliferative disorders, in which the detection of the mutation can aid in diagnosis and treatment. JAK2 is a cytoplasmic tyrosine kinase that participate in signaling pathways of cytokines. The mutation (thymine to guanine) which leads to a change of valine from phenylalanine turns JAK2 active and leads to rapid cell growth. Utilizing a procedure with ARMS has allowed researchers to create a diagnostic test for the disorder.
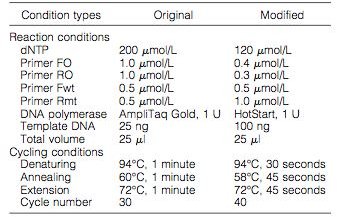
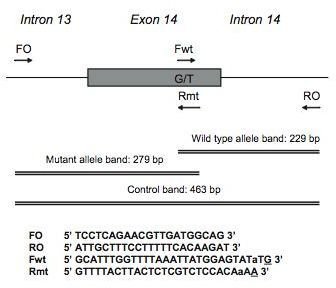
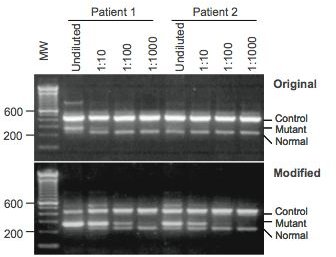
The researchers isolated DNA from multiple patients with human myeloproliferative disorders, and ran samples through an original and modified methods of ARMS-PCR (Figure 1). The design of the primers and the assay is found in Figure 2, which details where the primers attached to the JAK2 gene.
At the conclusion of the study, the researchers were able to use the modified version of ARMS for clinical testing of the JAK2 mutation that yielded better results. With the modified version, the gel electrophoresis gave clearer results than the original; this is due to the high sensitivity of ARMS (Figure 3). The results of the study gave a sensitivity range of about 0.05 to 0.1% making ARMS a helpful diagnostic tool in examining human myeloproliferative disorders.
Figures
Genotyping Measles Virus by RT-ARMS PCR
The measles virus (MV) infection is a large public heath problem with over 30 million people infected and around 530,000 deaths in 2003. Due to the large numbers and in an attempt to erradicate the disease by 2010, the World Health Organization recommends genotyping strains involved in outbreaks. The recommended procedure involves nucleotidic sequencing of genomic fragments followed by analysis. This method is not only expensive but is very time consuming, not allowing to be helpful in a time of an outbreak. A new method of real-time ARMS-PCR was demonstrated in a study and shown to have 97% concordance rate with direct sequencing.
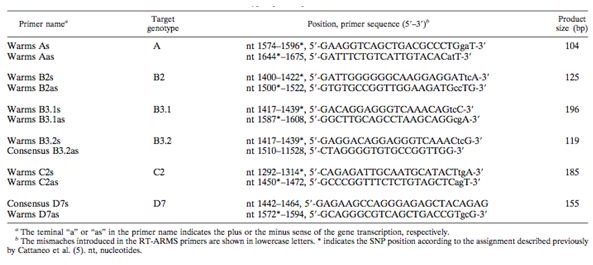
In the study, researches took 30 isolates from MV outbreaks and analyzed the samples with a method of RT-ARMS PCR along with SYBR green fluorescent dye. The technique allowed for genotype-specific primes to specifically amplify MV genotypes. The 450 C-terminal nucleotides of the N gene in MV contains enough nucleotide variations to allow discrimination among genotypes. This region was used in the development of an assay that would differentiate between different genotypes by using primers that bind to different SNPs. The primers that were designed to discrimnate between genotypes are found in Figure 4.
The results showed advantages over direct sequencing in rapidity, reproducibility and accuracy, which would be attractive for large scale-use. The results that were obtained by direct sequencing as a reference, and compared with the results obtained from the RT-ARMS PCR show that the accuracy of MV genotyping by melting curve analysis was 97%. However, despite similar results, RT-ARMS PCR held many advantages over direct sequencing.
- RT-ARMS PCR technique is more sensitive. Direct sequencing requires 50ng of PCR product whereas ARMS requires only .26ng.
- The time saved is notable. Direct sequencing for 30 samples requires around 3 days whereas ARMS needed only 2 hours.
- Lastly, ARMS is a more appropriate choice for large scale usage.
Why Is It Important?
Amplification refractory mutation system has a variety of uses in molecular genetics. It allows clinicians to test for mutational cause of sickle-cell anemia, chronic myeloproliferative disorders, examine new strains of the measles virus in outbreaks, and even find a diagnostic test for hemochromatosis. Even though ARMS is a similar in protocol to PCR, it is more accurate in genotyping. ARMS allows clinicians to not only test for a disorder, but even find the genotype of the patient undergoing an examination. With more experimentation of ARMS, researchers will find more uses for this technique, and hopefully unlock more diagnostic tools for other disorders. Using the technology is definitely an asset to the molecular genetic community.
References
- Amplification Refractory Mutation System Protocol. (n.d.). from https://www.ithanet.eu
- Chen, Q., Lu, P., Jones, A., Cross, N., Silver, R., & Wang, Y. (2007). Amplification
refractory mutation system, a highly sensitive and simple polymerase chain reaction
assay, for the detection of JAK2 V617F mutation in chronic myeloproliferative
disorders.. The Journal of Molecular Diagnostics , 9(2), 272-276. - Erlich, H., & Arnheim, N. (1992). Genetic Analysis Using The Polymerase Chain
Reaction. Annual Reviews of Genetics, 26, 479-506. - Newton, C., Graham, A., Heptinstall, L., Powell, S., Summers, C., Kalsheker, N.,
Smith, J., & Markham, A. (1989). Analysis of any point mutation in DNA. The
amplification refractory mutation system (ARMS). Nucl. Acids Res., 17(7),
2503-2516. - Pasternak, J. J. (2005). An Introduction to Human Molecular Genetics: Mechanisms
of Inherited Diseases. United States of America: Wiley-Liss. - Pissard, S., Huynh, L., Martin, J., & Goosens, M. (2002). HFE Genotyping by
Amplification Refractory Mutation System-Denaturing HPLC. Clinical Chemistry,
48(2), 769-772. - Waku-Kouomou, D., Alla, A., Blanquier, B., Jeantet, D., Caidi, H., Rguig, A.,
Freymuth, F., & Wild, F. (2006). Genotyping Measles Virus by Real-Time
Amplification Refractory Mutation System PCR Represents a Rapid Approach for
Measles Outbreak Investigations. Journal of Clinical Microbiolgy, 44(2), 487-494. - Logan, J., Edwards, K., Saunders, N. (2009). Real-Time PCR: Current Technology
and Applications. Wymondham: Caister Academic Press.